Mastering the Many Models Approach
A Comprehensive Guide to the Tidyverse Many Models Approach and its Extensions
Intro
The tidyverse “many models” approach was formally introduced in the first edition of R for Data Science (R4DS) in 2017. The central idea of this approach is to streamline the process of running the same model on various distinct subsets of data.
Since 2017, the tidyverse has evolved significantly, and along with it, the way we can harness the Many Models Approach. In this blog post we’ll first review the basic approach and update it using the latest tidyverse syntax. We’ll then explore a range of use cases with increasing complexity while introducing new building blocks and helper functions. This leads us finally up to the case where we not only run the same model on various, partly overlapping, subsets of our data, but where we also run slightly different models on those subsets simultaneously.
The structure of this blog post also reflects my motivation for writing it. I think that this is a powerful approach that should be more widely known. Those who are actually using it, often rely on an older syntax, which makes things more complicated than necessary. In addition to the original building blocks, there are several lesser-known functions that help apply this approach to more complex use cases.
Lately, the tidyverse Many Models Approach hasn’t received much attention. One might expect this to change with the release of the second edition of R4DS. However, the entire section on modeling has been omitted from this release. According to the authors, the reasons for this are twofold: First, there was never ample room to address the whole topic of modeling within R4DS. Second, the authors recommend the ‘tidymodels’ packages, which are well documented in Tidy Modeling with R and which should fill this gap.
While ‘tidymodels’ is a strong framework with definite advantages when working with various algorithms and model engines, it comes with considerable conceptual and syntactic overhead. For this reason, I believe there is still a lot room for the “classic” tidyverse Many Models Approach, which is based on ‘dplyr’ syntax but uses base R or alternatively package-specific models.
But before we delve into the use cases, let’s begin with the setup.
Setup
Unlike the name suggests, we don’t need all of the ‘tidyverse’ packages for the tidyverse Many Models Approach. The heavy lifting is done by ‘dplyr’ and ‘tidyr’. Additionally, we use ‘rlang’ and ‘purrr’ for some extra functionality. In this post we’ll be using the csat and csatraw data from my own package ‘dplyover’. It is important to note, that this post was written using ‘dplyr’ version 1.1.0. Readers who want to reproduce the code below should use at least ‘dplyr’ version version 1.0 or greater.
library(dplyr) # <- necessary
library(tidyr) # <- necessary
library(broom) # <- necessary
library(rlang) # <- nice to have
library(modelsummary) # <- for saving output
library(purrr) # <- not really needed
library(dplyover) # <- only for the datacsat is a mock-up dataset resembling data from a customer survey. It comes in two forms: the labeled data, csat, with meaningful column names and factor levels, and the corresponding raw data, csataw, where each column is a survey item and responses are just numbers. Since our models will be using the numbers instead of the factor levels, we’ll use the data from csatraw, but rename the columns according to csat (see my earlier post on how to rename variables). Additionally, we’ll drop variables that we don’t need. Let’s take a glimpse at the resulting dataset csat_named:
# create a look-up vector of old and new names
lookup_vec <- set_names(names(csatraw), names(csat))
# rename the columns
csat_named <- csatraw |>
rename(any_of(lookup_vec)) |>
select(cust_id, type, product, csat,
ends_with("rating"))
glimpse(csat_named)
#> Rows: 150
#> Columns: 9
#> $ cust_id <chr> "61297", "07545", "03822", "88219", "31831", "63646", "…
#> $ type <chr> "existing", "existing", "existing", "reactivate", "new"…
#> $ product <chr> "advanced", "advanced", "premium", "basic", "basic", "b…
#> $ csat <dbl> 3, 2, 2, 4, 4, 3, 1, 3, 3, 2, 5, 4, 4, 1, 4, 4, 2, 3, 2…
#> $ postal_rating <dbl> 3, 2, 5, 5, 2, NA, 3, 3, 4, NA, NA, 4, 3, 4, 1, 5, NA, …
#> $ phone_rating <dbl> 2, 4, 5, 3, 5, 3, 4, 2, NA, 2, NA, NA, 2, 4, NA, 2, 4, …
#> $ email_rating <dbl> NA, 3, NA, NA, 5, 2, 3, 5, 3, 1, 3, 1, 3, 1, 3, 3, 4, 1…
#> $ website_rating <dbl> 5, 2, 3, 4, 1, 3, 3, 1, 3, 2, 4, 2, NA, 4, 1, 2, NA, 5,…
#> $ shop_rating <dbl> 3, 1, 2, 2, 5, 4, 4, 2, 2, 2, 4, 3, 2, NA, 3, 5, 4, 1, …
Every row is the response of a customer, cust_id, who owns a contract-base product available in different flavors: “basic”, “advanced” or “premium”. There are three different types of customers: “existing”, “new” and “reactivate”. Our dependent variable is the customer satisfaction score, csat, which ranges from ‘1 = Very unsatisfied’ to ‘5 = Very satisfied’. The independent variables are ratings on the same scale concerning the following touchpoints: “postal”, “phone”, “email”, “website” and “shop”. We’ve dropped all other variables, but interested readers can find both datasets well documented in the ‘dplyover’ package (
?csat,
?csatraw).
Fundamentals
Let’s start with the basic approach as it was introduced in R4DS. Keep in mind that syntax and functions have evolved over the past five years, so we’ll be refining the original ideas into a more canonical form.
Before we dive into the details let’s look at the whole workflow from a computational perspective. We start by breaking down our original data into small grouped subsets, where each data set is stored in a row of a so called “nested” data.frame. The model fitting function is applied iteratively to each row. The model fits are appended to a new column of the nested data frame, again, one summary per row. We further process and tidy the results by turning the original model output into a list of small data.frames which can also be appended to a new column. Finally, we concatenate the small data.frames in each row and turn them into one big unified data.frame holding the results of all subgroups.
There are four essential components in the workflow above that we’ll be discussing in detail below:
- nested data
- rowwise operations
- tidy results
- unnesting results
If you’re already familiar with these concepts, feel free to skip this section.
Nested data
The central idea of the Many Models Approach is to streamline the process of running models on various subsets of data. Let’s say we want to perform a linear regression on each product type. In a traditional base R approach, we might have used a for loop to populate a list object with the results of each run. However, the tidyverse method begins with a nested data.frame.
So, what is a nested data.frame? We can use dplyr::nest_by(product) to create a data.frame containing three rows, one for each product. The second column, data, is a ‘list-column’ that holds a list of data.frame's—one for each row. These data.frames contain data for all customers within the corresponding product category. If you’re unfamiliar with list-columns, I highly recommend reading chapter 25 of R4DS. Although some parts may be outdated, it remains an excellent resource for understanding the essential components of this approach.
csat_prod_nested <- csat_named |>
nest_by(product)
csat_prod_nested
#> # A tibble: 3 × 2
#> # Rowwise: product
#> product data
#> <chr> <list<tibble[,8]>>
#> 1 advanced [40 × 8]
#> 2 basic [60 × 8]
#> 3 premium [50 × 8]
Looking at the first element (row) of the data column shows a data.frame with 40 customers, all of whom have “advanced” products. The product column is omitted, as this information is already included in our nested data: csat_prod_nested.
csat_prod_nested$data[[1]]
#> # A tibble: 40 × 8
#> cust_id type csat postal_rating phone_rating email_r…¹ websi…² shop_…³
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 61297 existing 3 3 2 NA 5 3
#> 2 07545 existing 2 2 4 3 2 1
#> 3 63600 existing 1 3 4 3 3 4
#> 4 82048 reactivate 3 3 2 5 1 2
#> 5 41142 reactivate 3 4 NA 3 3 2
#> 6 06387 reactivate 1 4 4 1 4 NA
#> 7 63024 existing 2 NA 4 4 NA 4
#> 8 55743 new 1 4 5 4 1 5
#> 9 32689 new 5 3 NA 3 4 2
#> 10 33603 existing 2 3 3 4 3 2
#> # … with 30 more rows, and abbreviated variable names ¹email_rating,
#> # ²website_rating, ³shop_rating
Rowwise operations
Applying
nest_by() also groups our data
rowwise(). This means that subsequent dplyr operations will be applied “one row at a time.” This is particularly helpful when vectorized functions aren’t available, such as the
lm() function in our case, which we want to apply to the data in each row.
First, let’s define the relationship between our dependent and independent variables using a formula object, base_formula.
base_formula <- csat ~ postal_rating + phone_rating + email_rating +
website_rating + shop_rating
base_formula
#> csat ~ postal_rating + phone_rating + email_rating + website_rating +
#> shop_rating
Note, that this is a basic “model formula” which holds the dependent variable, csat, on the left-hand side (of the tilde ~) and all independent variables on the right-hand side.
Next, we use mutate to create new columns. We’ll start by creating a column called mod containing our model. We’ll apply the
lm() function with the previously defined formula and supply the data column to it. Note, that when supplying arguments within
dplyr::mutate() we can use both: (i) objects from the data.frame we are working on, here the data column of our csat_prod_nested dataset, and (ii) objects from the enclosing environment, here the global environment, where our formula object base_formula resides.
Since we are working with a rowwise data.frame, the
lm() function is executed three times, one time for each row, each time using a different data.frame in the list-column data. As the result of each call is not an atomic vector, but an lm object of type list, we need to wrap the function call in
list(). This results in a new list-column, mod, which holds an lm object in each row.
csat_prod_nested |>
mutate(mod = list(lm(base_formula, data = data)))
#> # A tibble: 3 × 3
#> # Rowwise: product
#> product data mod
#> <chr> <list<tibble[,8]>> <list>
#> 1 advanced [40 × 8] <lm>
#> 2 basic [60 × 8] <lm>
#> 3 premium [50 × 8] <lm>
Two notes of caution when using
dplyr::rowwise():
First, keep in mind that we need the wrapping
list() call whenever the result is not an atomic vector. Since this is fairly often the case, it’s good practice to resonate about the return type of a function when using
dplyr::rowwise().
Second, once we call
rowwise() on a
data.frame(), all following operations will be applied row-by-row. We can revoke this by calling
ungroup() on a rowwise data.frame. Sometimes we might forget to
ungroup(). In most cases this will not lead to an error. However, it will lead to a degraded performance, since vectorized functions, which are supposed to work on whole columns, are now unnecessarily applied row-by-row.
Tidy results with broom
To make the results of this model more accessible, we’ll use two functions from the ‘broom’ package:
broom::glance() returns a data.frame containing all model statics, such as r-squared, BIC, AIC etc., and the overall p-value of the model itself.
broom::tidy() returns a data.frame with all regression terms, their estimates, p-values and other statistics.
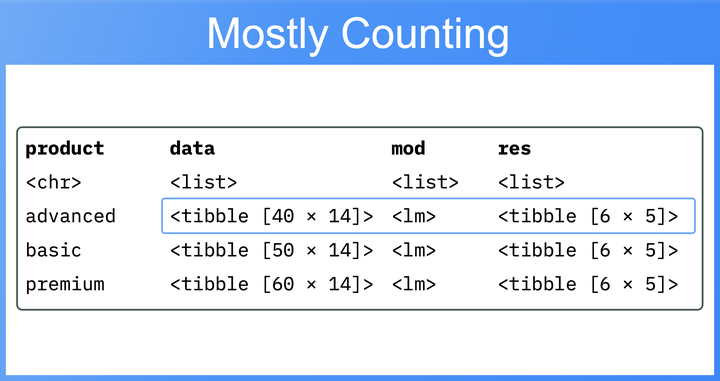
Again, we’ll wrap both functions in
list() and call them on the model in the new mod column. This yields a final, nested data.frame. The rows represent the three product subgroups, while the columns contain the input data, the model mod, and the results modstat and res. Beside mod, each of these columns is a list of data.frames:
csat_prod_nested_res <- csat_prod_nested |>
mutate(mod = list(lm(base_formula, data = data)),
modstat = list(broom::glance(mod)),
res = list(broom::tidy(mod)))
csat_prod_nested_res
#> # A tibble: 3 × 5
#> # Rowwise: product
#> product data mod modstat res
#> <chr> <list<tibble[,8]>> <list> <list> <list>
#> 1 advanced [40 × 8] <lm> <tibble [1 × 12]> <tibble [6 × 5]>
#> 2 basic [60 × 8] <lm> <tibble [1 × 12]> <tibble [6 × 5]>
#> 3 premium [50 × 8] <lm> <tibble [1 × 12]> <tibble [6 × 5]>
Unnesting results
With the groundwork laid, it is now easy to access the results. To do this, we’ll use
tidyr::unnest() to convert a list of data.frames back into a regular data.frame. First, lets look at the model statistics. We’ll select the product and modstat columns and unnest the latter. This produces a data.frame with different model statistics for the three product subgroups. In this case, we’re interested in the r-squared, the p-value and the number of observations of each model:
csat_prod_nested_res |>
select(product, modstat) |>
unnest(modstat) |>
select(r.squared, p.value, nobs)
#> Adding missing grouping variables: `product`
#> # A tibble: 3 × 4
#> # Groups: product [3]
#> product r.squared p.value nobs
#> <chr> <dbl> <dbl> <int>
#> 1 advanced 0.112 0.941 15
#> 2 basic 0.382 0.192 20
#> 3 premium 0.185 0.645 21
Please note that the results themselves aren’t the main focus here, as the primary goal is to demonstrate how the approach works in general.
Next, we’ll inspect the coefficients, their size and their p-values. We’ll select the product and res columns and unnest the latter. Additionally, since we’re not interested in the size of the intercept, we’ll filter out those rows.
csat_prod_nested_res |>
select(product, res) |>
unnest(res) |>
filter(term != "(Intercept)")
#> # A tibble: 15 × 6
#> # Groups: product [3]
#> product term estimate std.error statistic p.value
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 advanced postal_rating 0.396 0.483 0.819 0.434
#> 2 advanced phone_rating -0.235 0.423 -0.556 0.592
#> 3 advanced email_rating 0.349 0.485 0.720 0.490
#> 4 advanced website_rating 0.398 0.456 0.874 0.405
#> 5 advanced shop_rating -0.00183 0.288 -0.00637 0.995
#> 6 basic postal_rating -0.324 0.225 -1.44 0.172
#> 7 basic phone_rating -0.297 0.237 -1.25 0.231
#> 8 basic email_rating -0.136 0.249 -0.547 0.593
#> 9 basic website_rating 0.229 0.229 1.00 0.334
#> 10 basic shop_rating 0.321 0.259 1.24 0.235
#> 11 premium postal_rating 0.0538 0.220 0.245 0.810
#> 12 premium phone_rating -0.508 0.307 -1.65 0.119
#> 13 premium email_rating 0.375 0.309 1.22 0.243
#> 14 premium website_rating -0.175 0.259 -0.677 0.509
#> 15 premium shop_rating 0.0498 0.203 0.246 0.809
From this point, we could further manipulate the resulting data, such as filtering out non-significant coefficients or plotting the model results, and so on.
To wrap up this section, the info box below highlights how the approach above deviates from the original syntax introduced in R4DS.
Expand: The original syntax
There are two main differences between the approach outlined above and the syntax which was originally introduced in R4DS.
First, instead of nest_by(product), the original syntax used group_by(product) %>% nest(). Both produce a nested data.frame. The later, however, returns a data.frame grouped by “product”, while
nest_by() returns a rowwise data.frame.
While this difference seems negligible, it at has implications on how operations on the nested data are carried out, especially, since rowwise operations didn’t exist in 2017. The original approach was using
purrr::map() and friends instead to apply unvectorized functions, such as
lm(), to list-columns.
csat_named |>
group_by(product) |>
nest() |>
ungroup() |>
mutate(mod = map(data, ~ lm(base_formula, data = .x)),
res = map(mod, broom::tidy),
modstat = map(mod, broom::glance))
#> # A tibble: 3 × 5
#> product data mod res modstat
#> <chr> <list> <list> <list> <list>
#> 1 advanced <tibble [40 × 8]> <lm> <tibble [6 × 5]> <tibble [1 × 12]>
#> 2 premium <tibble [50 × 8]> <lm> <tibble [6 × 5]> <tibble [1 × 12]>
#> 3 basic <tibble [60 × 8]> <lm> <tibble [6 × 5]> <tibble [1 × 12]>
While this approach saves us from wrapping the output in
list(), it leads to code cluttering especially with functions that take two or more arguments and which need to be wrapped in
pmap().
Extensions
Building on the basic approach outlined above, we’ll introduce six advanced building blocks that help to tackle more complex use cases.
- create an overall category with
bind_rows() - add subgroups through filters with
expand_grid() - dynamically name list elements with
rlang::list2() - use data-less grids
- build formulas programmatically with
reformulate() - save model output to Excel with
modelsummary()
Create an overall category with ‘bind_rows()’
Often we want to run an analysis not only on different subsets of the data but also on the entire dataset simultaneously. We can achieve this by using
mutate() and
bind_rows() to create an additional overall product category that encompasses all products.
csat_all <- csat_named |>
mutate(product = "All") |>
bind_rows(csat_named)
csat_all |> count(product)
#> # A tibble: 4 × 2
#> product n
#> <chr> <int>
#> 1 All 150
#> 2 advanced 40
#> 3 basic 60
#> 4 premium 50
First, we use
mutate() to overwrite the ‘product’ column with the value “All” effectively grouping all products together under this new label. We then use
bind_rows() to merge the original csat_named dataset with the modified dataset from the previous step. This results in a new dataset called csat_all that contains the original data and an extra set of rows where the product category is labeled as “All”. Consequently, the new dataset is twice the size of the original data, as it includes each row twice.
Now we can apply the same analysis as above:
csat_all |>
nest_by(product) |>
mutate(mod = list(lm(base_formula, data = data)),
res = list(broom::tidy(mod)),
modstat = list(broom::glance(mod)))Expand to show output
#> # A tibble: 4 × 5
#> # Rowwise: product
#> product data mod res modstat
#> <chr> <list<tibble[,8]>> <list> <list> <list>
#> 1 All [150 × 8] <lm> <tibble [6 × 5]> <tibble [1 × 12]>
#> 2 advanced [40 × 8] <lm> <tibble [6 × 5]> <tibble [1 × 12]>
#> 3 basic [60 × 8] <lm> <tibble [6 × 5]> <tibble [1 × 12]>
#> 4 premium [50 × 8] <lm> <tibble [6 × 5]> <tibble [1 × 12]>
Add subgroups through filters with ‘expand_grid()’
Sometimes, we may want to create additional subgroups that meet specific filter criteria. For instance, we might want to analyze all customers and, at the same time, compare the results with an analysis of all customers who are not of the “reactivate” type.
To achieve this, we’ll follow three steps:
-
We create a list of filter expressions, referred to as
filter_ls.filter_ls <- list( All = TRUE, no_reactivate = expr(type != "reactivate") ) filter_ls #> $All #> [1] TRUE #> #> $no_reactivate #> type != "reactivate"This results in a list where each element is either
TRUEor an unevaluated expression that we’ll use later insidefilter(). Note that we userlang::expr()to capture an expression, although we could have usedsubstitute()orquote()instead. Omitting any of these function would result in an error, as R would try to evaluatetypewhich is not desired since we want to delay the evaluation until later. -
We expand our nested
data.framefor each filter category usingexpand_grid().csat_all_grps <- csat_all |> nest_by(product) |> expand_grid(filter_ls) |> mutate(type = names(filter_ls), .after = product) csat_all_grps #> # A tibble: 8 × 4 #> product type data filter_ls #> <chr> <chr> <list<tibble[,8]>> <named list> #> 1 All All [150 × 8] <lgl [1]> #> 2 All no_reactivate [150 × 8] <language> #> 3 advanced All [40 × 8] <lgl [1]> #> 4 advanced no_reactivate [40 × 8] <language> #> 5 basic All [60 × 8] <lgl [1]> #> 6 basic no_reactivate [60 × 8] <language> #> 7 premium All [50 × 8] <lgl [1]> #> 8 premium no_reactivate [50 × 8] <language>We use
tidyr::expand_grid()to expand our nested data for each category in our list of filter expressions:filter_ls.expand_grid()creates atibblefrom all combinations of the supplied objects—this can be any mix of atomic vectors, lists ordata.frames. Note that in the case of the latter, all groupings are ignored. Looking at the output reveals that our original nesteddata.framecontained four rows, while our data now holds eight rows - one for each combination ofproductandtype.We also use
dplyr::mutate()to add a new column,type, to actually show which row refers to which type. Thenames()of our list columnfilter_lscontain this information. To show thetypenext to theproductcolumn we use the argument.afterinmutate(). Without specifying this, thetypecolumn would have been added as the last column in our nesteddata.frame. -
We apply each filter to our data
rowwiseusingdplyr::filter(eval(filter_ls)).csat_all_grps_grid <- csat_all_grps |> rowwise() |> mutate(data = list( filter(data, eval(filter_ls)) ), .keep = "unused" ) csat_all_grps_grid #> # A tibble: 8 × 3 #> # Rowwise: #> product type data #> <chr> <chr> <list> #> 1 All All <tibble [150 × 8]> #> 2 All no_reactivate <tibble [120 × 8]> #> 3 advanced All <tibble [40 × 8]> #> 4 advanced no_reactivate <tibble [32 × 8]> #> 5 basic All <tibble [60 × 8]> #> 6 basic no_reactivate <tibble [46 × 8]> #> 7 premium All <tibble [50 × 8]> #> 8 premium no_reactivate <tibble [42 × 8]>We use
mutateto applyfilter()to eachdata.framein thedatacolumn in each row. As filter expression we use the columnfilter_lsand evaluate it. Since we no longer need this column, we set the.keepargument inmutateto “unused” to eventually dropfilter_lsafter it has been used.
From this point, we could continue applying our model and then calculating and extracting the results, but we’ll omit this for the sake of brevity.
Expand to show code
csat_all_grps_grid <- csat_all_grps |>
rowwise() |>
mutate(mod = list(lm(base_formula, data = data)),
res = list(broom::tidy(mod)),
modstat = list(broom::glance(mod)))
csat_all_grps_grid |>
select(product, type, modstat) |>
unnest(modstat) |>
select(-c(sigma, statistic, df:df.residual))
#> # A tibble: 8 × 6
#> product type r.squared adj.r.squared p.value nobs
#> <chr> <chr> <dbl> <dbl> <dbl> <int>
#> 1 All All 0.134 0.0479 0.191 56
#> 2 All no_reactivate 0.134 0.0479 0.191 56
#> 3 advanced All 0.112 -0.381 0.941 15
#> 4 advanced no_reactivate 0.112 -0.381 0.941 15
#> 5 basic All 0.382 0.161 0.192 20
#> 6 basic no_reactivate 0.382 0.161 0.192 20
#> 7 premium All 0.185 -0.0867 0.645 21
#> 8 premium no_reactivate 0.185 -0.0867 0.645 21
csat_all_grps_grid |>
select(product, type, res) |>
unnest(res) |>
filter(term == "website_rating")
#> # A tibble: 8 × 7
#> product type term estimate std.error statistic p.value
#> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 All All website_rating 0.0729 0.141 0.517 0.607
#> 2 All no_reactivate website_rating 0.0729 0.141 0.517 0.607
#> 3 advanced All website_rating 0.398 0.456 0.874 0.405
#> 4 advanced no_reactivate website_rating 0.398 0.456 0.874 0.405
#> 5 basic All website_rating 0.229 0.229 1.00 0.334
#> 6 basic no_reactivate website_rating 0.229 0.229 1.00 0.334
#> 7 premium All website_rating -0.175 0.259 -0.677 0.509
#> 8 premium no_reactivate website_rating -0.175 0.259 -0.677 0.509
Although this example is relatively simple, it demonstrates how this approach can be significantly expanded by providing more filter expressions in our filter_ls list or by using multiple lists of filter expressions. This is particularly useful when performing robustness checks, where we attempt to reproduce the original findings on specific subgroups of our data.
Dynamically name list elements with ‘rlang::list2()’
So far, we’ve wrapped the results of our rowwise operations in
list() when they produced non-atomic vectors.
A common issue when inspecting the results is that these list-columns are often unnamed, making it difficult to determine which element we’re examining. For instance, suppose that we want to double-check the output of our call to broom::glance(mod) stored in the modstat column. Let’s look at the fourth element:
csat_all_grps_grid$modstat[4]
#> [[1]]
#> # A tibble: 1 × 12
#> r.squ…¹ adj.r…² sigma stati…³ p.value df logLik AIC BIC devia…⁴ df.re…⁵
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int>
#> 1 0.112 -0.381 1.39 0.228 0.941 5 -22.4 58.9 63.8 17.5 9
#> # … with 1 more variable: nobs <int>, and abbreviated variable names
#> # ¹r.squared, ²adj.r.squared, ³statistic, ⁴deviance, ⁵df.residual
The result prints nicely, but it’s unclear which subset of the data it belongs to.
Here
rlang::list2() comes to the rescue. Although it resembles
list(), it provides some extra functionality. Specifically, it allows us to unquote names on the left-hand side of the walrus operator. To better grasp this idea, let’s look at an example.
We wrap our calls to
lm(),
tidy() and
glance() in
list2() and name each element using the walrus operator :=. On the left-hand side of the walrus operator, we use the glue operator { within a string to dynamically name each element according to the values in the product and type columns in each row. When we inspect the fourth element of the modstat column, we can quickly see that these model statistics belong to the subset of customers with an “advanced” product and who are not of type “reactivate”.
csat_all_grps_grid <- csat_all_grps |>
rowwise() |>
mutate(mod = list2("{product}_{type}" := lm(base_formula, data = data)),
res = list2("{product}_{type}" := broom::tidy(mod)),
modstat = list2("{product}_{type}" := broom::glance(mod)))
csat_all_grps_grid$modstat[4]
#> $advanced_no_reactivate
#> # A tibble: 1 × 12
#> r.squ…¹ adj.r…² sigma stati…³ p.value df logLik AIC BIC devia…⁴ df.re…⁵
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int>
#> 1 0.112 -0.381 1.39 0.228 0.941 5 -22.4 58.9 63.8 17.5 9
#> # … with 1 more variable: nobs <int>, and abbreviated variable names
#> # ¹r.squared, ²adj.r.squared, ³statistic, ⁴deviance, ⁵df.residual
Data-less grids
Using the methods described above, we can easily construct nested data.frames with several dozens of subgroups. However, this approach can be inefficient in terms of memory usage, as we create a copy of our data for every single subgroup. To make this approach more memory-efficient, we can use what I call a “data-less grid”, which is similar to our original nested data.frame, but without the data column.
Instead of nesting our data with
nest_by(), we manually create the combinations of subgroups to which we want to apply our model. We start with a vector of all unique values in the product column and add an overall category “All” to it. Then, we supply this vector along with our list of filter expressions filter_ls to
expand_grid(). Finally, we place the names of the elements in filter_ls in a separate column: type.
This results in an initial grid all_grps_grid of combinations between product and type, with an additional column of filter expressions.
product <- c(
"All", unique(csat_named$product)
)
all_grps_grid <- expand_grid(product, filter_ls) |>
mutate(type = names(filter_ls),
.after = product)
all_grps_grid
#> # A tibble: 8 × 3
#> product type filter_ls
#> <chr> <chr> <named list>
#> 1 All All <lgl [1]>
#> 2 All no_reactivate <language>
#> 3 advanced All <lgl [1]>
#> 4 advanced no_reactivate <language>
#> 5 premium All <lgl [1]>
#> 6 premium no_reactivate <language>
#> 7 basic All <lgl [1]>
#> 8 basic no_reactivate <language>
The challenging aspect here is to generate each data subset on the fly in the call to
lm(). To accomplish this, we
filter() our initial data csat_named on two conditions:
-
Firstly, we filter for different
producttypes using an advanced filter expression:.env$product == "All" | .env$product == product.This expression may appear somewhat obscure, so let’s break it down:
The issue here is that both our original data
csat_namedand our gridall_grps_gridcontain a column namedproduct. By default,product, in thefilter()call below, refers to the column incsat_named. To tell ‘dyplr’ to use the column in our gridall_grps_gridwe use the.envpronoun.So, the filter expression above essentially states: If the product category in our grid
.env$productis “All”, then select all rows. This works because when the left side of the or-condition.env$product == "All"evaluates toTRUE,filterselects all rows. If the first part of our condition is not true, then theproductcolumn incsat_namedshould match the value of theproductcolumn of our data-less grid.env$product. -
Next, we filter the different
types of customers. Here, we use the filter expressions stored infilter_lsand evaluate them. Since we don’t need thefilter_lscolumn after we’ve used it to create thedatacolumn, we just drop it withselect(! filter_ls):
all_grps_grid_mod <- all_grps_grid |>
rowwise() |>
mutate(mod = list(
lm(base_formula,
data = filter(csat_named,
# 1. filter product categories
.env$product == "All" | .env$product == product,
# 2. filter customer types
eval(filter_ls)
)
)
)
) |>
select(! filter_ls)
all_grps_grid_mod
#> # A tibble: 8 × 3
#> # Rowwise:
#> product type mod
#> <chr> <chr> <list>
#> 1 All All <lm>
#> 2 All no_reactivate <lm>
#> 3 advanced All <lm>
#> 4 advanced no_reactivate <lm>
#> 5 premium All <lm>
#> 6 premium no_reactivate <lm>
#> 7 basic All <lm>
#> 8 basic no_reactivate <lm>
This returns our initial grid, now extended by an additional column, mod, containing the linear models.
The remaining steps do not significantly differ from our initial approach, so we will omit them for brevity.
Expand to show code
all_grps_grid_res <- all_grps_grid_mod |>
mutate(res = list(broom::tidy(mod)),
modstat = list(broom::glance(mod)))
all_grps_grid_res |>
select(product, type, modstat) |>
unnest(modstat) |>
select(-c(sigma, statistic, df:df.residual))
#> # A tibble: 8 × 6
#> product type r.squared adj.r.squared p.value nobs
#> <chr> <chr> <dbl> <dbl> <dbl> <int>
#> 1 All All 0.134 0.0479 0.191 56
#> 2 All no_reactivate 0.180 0.0771 0.145 46
#> 3 advanced All 0.112 -0.381 0.941 15
#> 4 advanced no_reactivate 0.330 -0.340 0.772 11
#> 5 premium All 0.185 -0.0867 0.645 21
#> 6 premium no_reactivate 0.156 -0.196 0.810 18
#> 7 basic All 0.382 0.161 0.192 20
#> 8 basic no_reactivate 0.465 0.221 0.172 17
Build formulas programmatically with ‘reformulate’
One last major building block that essentially completes the Many Models Approach is actually a base R function:
reformulate().
I recently posted an #RStats meme on Twitter highlighting that
reformulate() is one of the lesser-known base R functions, even among advanced users. The reactions to my post largely confirmed my impression.
Here you'll find additional content from Twitter that complements the article. You can easily view it with a single click and then hide it again. I agree to have external content displayed to me. This may result in personal data being shared with third-party platforms. To learn more, please refer to our privacy policy.External Content
Before applying
reformulate() in the many models context, let’s have a look at what it does. Instead of manually creating a formula object by typing y ~ x1 + x2, we can use
reformulate() to generate a formula object based on character vectors.
Important is the order of the first two arguments. While we start writing a formula from the left-hand side y,
reformulate() takes as first argument the right-hand side. We could, of course, change the order and call the arguments by name instead of position, but I wouldn’t recommend this, because it might confuse readers. Nevertheless, calling the arguments by name, termlabels and response, and their correct position is a good idea to make clear which is which and to remind readers that the argument order is different to what they might expect.
form1 <- y ~ x1 + x2
form1
#> y ~ x1 + x2
form2 <- reformulate(termlabels = c("x1", "x2"),
response = "y")
form2
#> y ~ x1 + x2
identical(form1, form2)
#> [1] TRUE
How can we make use of
reformulate() in the Many Models Approach?
In the following we’ll look at two use cases. The first use case covers a situation where we need to fit multiple models to the data in each row of a nested data frame. All of these models share the same response variable and we use
reformulate() to supply different predictor variables in each iteration. The second use case covers a situation where we also need to fit multiple models to the data in each row of a nested data frame. However, this time we define a base model in advance and add different variables on top of it in each iteration.
Let’s begin with the first use case outline above in which we want to construct a separate model for each independent variable, containing only our response variable and one independent variable at a time: csat ~ indepedent_variable. And of course, we want to do this for all of our subgroups of the previous approach.
First, we need a character vector holding the names of our independent variables. With this vector, we can now expand our data-less grid from above. This results in a new grid with 40 rows (eight subgroups times five independent variables).
indep_vars <- c("postal_rating",
"phone_rating",
"email_rating",
"website_rating",
"shop_rating")
all_grps_grid_vars <- all_grps_grid |>
expand_grid(indep_vars)
all_grps_grid_vars
#> # A tibble: 40 × 4
#> product type filter_ls indep_vars
#> <chr> <chr> <named list> <chr>
#> 1 All All <lgl [1]> postal_rating
#> 2 All All <lgl [1]> phone_rating
#> 3 All All <lgl [1]> email_rating
#> 4 All All <lgl [1]> website_rating
#> 5 All All <lgl [1]> shop_rating
#> 6 All no_reactivate <language> postal_rating
#> 7 All no_reactivate <language> phone_rating
#> 8 All no_reactivate <language> email_rating
#> 9 All no_reactivate <language> website_rating
#> 10 All no_reactivate <language> shop_rating
#> # … with 30 more rows
We can now apply a similar approach as before, creating data subgroups on the fly. The only change is that we use reformulate(indep_vars, "csat") instead of our formula object base_formula. This adds forty different linear models to our grid:
all_grps_grid_vars_mod <- all_grps_grid_vars |>
rowwise() |>
mutate(mod = list(
lm(reformulate(indep_vars, "csat"), # <- this part is new
data = filter(csat_named,
.env$product == "All" | .env$product == product,
eval(filter_ls)
)
)
)
) %>%
select(! filter_ls)
all_grps_grid_vars_mod
#> # A tibble: 40 × 4
#> # Rowwise:
#> product type indep_vars mod
#> <chr> <chr> <chr> <list>
#> 1 All All postal_rating <lm>
#> 2 All All phone_rating <lm>
#> 3 All All email_rating <lm>
#> 4 All All website_rating <lm>
#> 5 All All shop_rating <lm>
#> 6 All no_reactivate postal_rating <lm>
#> 7 All no_reactivate phone_rating <lm>
#> 8 All no_reactivate email_rating <lm>
#> 9 All no_reactivate website_rating <lm>
#> 10 All no_reactivate shop_rating <lm>
#> # … with 30 more rows
Note, that we again drop the filter_ls column after we used it to create out data.
Although the example above is instructive, it isn’t particularly useful. In most cases, we don’t want to create a separate model for each independent variable. A much more powerful way to use
reformulate() is to
update() a baseline model with additional variables. So let’s look at our second use case.
Assume we have the following minimal version of our base-line model:
min_formula <- csat ~ postal_rating + phone_rating + shop_rating
min_formula
#> csat ~ postal_rating + phone_rating + shop_rating
For our many subgroups from above, we want to check if adding (i) email_rating or (ii) website_rating or (iii) both variables at once, or (iv) both variables and their interaction effect, improve our model. Let’s create a list of terms that we want to add to our model: update_vars.
update_vars <- list(base = NULL,
email = "email_rating",
website = "website_rating",
both = c("email_rating", "website_rating"),
both_plus_interaction = "email_rating*website_rating")There are two noteworthy aspects here. First, we need to include NULL, as this will represent our baseline model without any additional variables. Second, we are not limited to plain variable names as strings. We can even include interaction effects using the same notation that the tilde operator supports, that is by adding an asterisk * between two variable names.
Now we expand our grid from above with this list and put the names of each variable (“base”, “email”, “website” etc.) in a separate column to keep track of which model we are examining.
all_grid_upd_vars <- all_grps_grid |>
expand_grid(update_vars) |>
mutate(model_spec = names(update_vars),
.after = type)
all_grid_upd_vars
#> # A tibble: 40 × 5
#> product type model_spec filter_ls update_vars
#> <chr> <chr> <chr> <named list> <named list>
#> 1 All All base <lgl [1]> <NULL>
#> 2 All All email <lgl [1]> <chr [1]>
#> 3 All All website <lgl [1]> <chr [1]>
#> 4 All All both <lgl [1]> <chr [2]>
#> 5 All All both_plus_interaction <lgl [1]> <chr [1]>
#> 6 All no_reactivate base <language> <NULL>
#> 7 All no_reactivate email <language> <chr [1]>
#> 8 All no_reactivate website <language> <chr [1]>
#> 9 All no_reactivate both <language> <chr [2]>
#> 10 All no_reactivate both_plus_interaction <language> <chr [1]>
#> # … with 30 more rows
We could use
update() directly in our call to
lm(), but to avoid overcomplicating things, let’s create a column holding our updated formula, form, and use that in our call to
lm():
all_grid_upd_vars_form <- all_grid_upd_vars |>
rowwise() |>
mutate(
form = list(
update(
min_formula, # old formula
reformulate(c(".", update_vars)) # changes to formula
)
),
mod = list2( "{product}_{type}_{model_spec}" :=
lm(form,
data = filter(csat_named,
.env$product == "All" | .env$product == product,
eval(filter_ls)
)
)
)
)
update() takes two arguments, the formula we want to update, in this case min_formula, and the formula we use to update the former. In our case, this is a call to
reformulate() which says: “take all the original term labels ".", and add
c() to them the variable in update_vars. Now its probably clear why we included NULL in update_vars. In cases where it is NULL the original formula won’t be updated, which corresponds to our baseline model.
Checking the first five rows of our list-column containing the model shows that the approach works as intended:
head(all_grid_upd_vars_form$mod, 5)Expand to show code
#> $All_All_base
#>
#> Call:
#> lm(formula = form, data = filter(csat_named, .env$product ==
#> "All" | .env$product == product, eval(filter_ls)))
#>
#> Coefficients:
#> (Intercept) postal_rating phone_rating shop_rating
#> 4.08357 0.02305 -0.26742 -0.11736
#>
#>
#> $All_All_email
#>
#> Call:
#> lm(formula = form, data = filter(csat_named, .env$product ==
#> "All" | .env$product == product, eval(filter_ls)))
#>
#> Coefficients:
#> (Intercept) postal_rating phone_rating shop_rating email_rating
#> 4.21064 -0.01306 -0.35218 -0.01432 0.01203
#>
#>
#> $All_All_website
#>
#> Call:
#> lm(formula = form, data = filter(csat_named, .env$product ==
#> "All" | .env$product == product, eval(filter_ls)))
#>
#> Coefficients:
#> (Intercept) postal_rating phone_rating shop_rating website_rating
#> 3.59965 -0.03583 -0.22622 -0.04578 0.13008
#>
#>
#> $All_All_both
#>
#> Call:
#> lm(formula = form, data = filter(csat_named, .env$product ==
#> "All" | .env$product == product, eval(filter_ls)))
#>
#> Coefficients:
#> (Intercept) postal_rating phone_rating shop_rating email_rating
#> 3.72401 -0.05946 -0.36126 0.05673 0.10789
#> website_rating
#> 0.07286
#>
#>
#> $All_All_both_plus_interaction
#>
#> Call:
#> lm(formula = form, data = filter(csat_named, .env$product ==
#> "All" | .env$product == product, eval(filter_ls)))
#>
#> Coefficients:
#> (Intercept) postal_rating
#> 3.46609 -0.05549
#> phone_rating shop_rating
#> -0.36445 0.05816
#> email_rating website_rating
#> 0.18616 0.15469
#> email_rating:website_rating
#> -0.02658
Save model output to Excel with ‘modelsummary()’
Although we previously used the ‘broom’ package to create tidy data.frames containing the model statistics, modstat created with
broom::glance(), and the model results, res created with
broom::tidy(), we ideally need both pieces of information when exporting the model output to Excel (or any other spreadsheet).
In this case, the
modelsummary() function from the package of the same name proves extremely helpful. It creates an Excel file that includes both model statistics and estimator results, which is convenient when reporting our model findings to a non-R-user audience.
The great feature of
modelsummary() is that it accepts list-columns of model objects, such as our mod column containing many lm objects, as input. We can specify various output formats—below we choose ".xlsx". Numerous other arguments allow us to trim the results for a more compact table. Here, we opt to omit the AIC, BIC, RMSE and log likelihood model statistics, as well as the coefficient size of the intercept. Setting the stars argument to TRUE adds the typical p-value stars to the estimators.
# this saves the results to a `data.frame` in `out` and ...
# at the same time creates an .xlsx file
out <- modelsummary(models = all_grid_upd_vars_form$mod,
output = "model_results.xlsx",
gof_omit = "AIC|BIC|Log.Lik|RMSE",
coef_omit = "(Intercept)",
stars = TRUE,
statistic = NULL)The following screenshot shows the resulting Excel table:
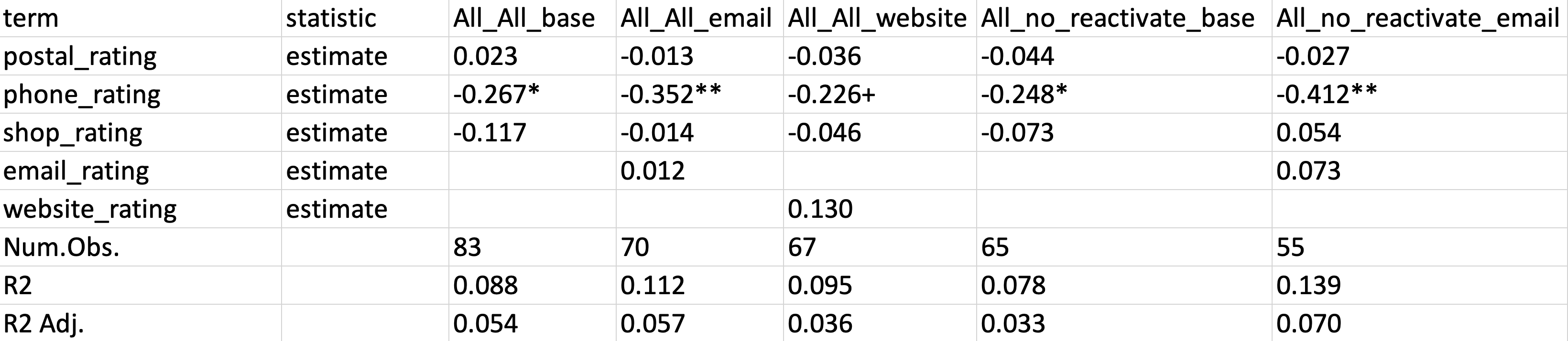
Examining the first few columns shows that not only the results print nicely, but they also include model names indicating the subgroups of each
lm() call. Accepting a named list-columns of model objects is indeed a fantastic feature of the
modelsummary() function.
The call to
modelsummary() above gives us a quick and compact overview of our results in Excel. However, one minor issue is that the p-value stars appear in the same column as the model coefficients. For a better presentation and reporting, I prefer having the stars in a separate column. To achieve this, we can set the output argument to "data.frame", add the stars as statistic, convert the results to long format with
pivot_wider() and save the resulting data.frame to Excel with
openxlsx::write.xlsx(). Since this is a minor issue, I leave the code for the interested reader in the output box below.
Expand to show code
out <- modelsummary(models = all_grid_upd_vars_form$mod,
output = "data.frame",
gof_omit = "AIC|BIC|Log.Lik|RMSE",
coef_omit = "(Intercept)",
statistic = "stars") |>
mutate(statistic = ifelse(statistic == "", "estimate", statistic)) |>
select(-part) |>
pivot_wider(names_from = statistic,
values_from = -c(term, statistic),
values_fn = as.numeric)
openxlsx::write.xlsx(out, "model_results2.xlsx")Endgame
With the building blocks introduced above, we can now combine everything and extend this approach even further.
Let’s say we want to compare two different versions of our dependent variable. The original variable and a collapsed top-box version csat_top taking 1 if a customer gave the best rating and 0 otherwise.
Again, we create a character vector holding the names of our dependent variables, dep_vars, and use it to expand our data-less grid from above.
csat_named_top <- csat_named |>
mutate(csat_top = ifelse(csat == 5, 1, 0))
dep_vars <- c("csat", "csat_top")
all_grps_grid_final <- all_grid_upd_vars |>
expand_grid(dep_vars)
all_grps_grid_final
#> # A tibble: 80 × 6
#> product type model_spec filter_ls update_vars dep_vars
#> <chr> <chr> <chr> <named list> <named list> <chr>
#> 1 All All base <lgl [1]> <NULL> csat
#> 2 All All base <lgl [1]> <NULL> csat_top
#> 3 All All email <lgl [1]> <chr [1]> csat
#> 4 All All email <lgl [1]> <chr [1]> csat_top
#> 5 All All website <lgl [1]> <chr [1]> csat
#> 6 All All website <lgl [1]> <chr [1]> csat_top
#> 7 All All both <lgl [1]> <chr [2]> csat
#> 8 All All both <lgl [1]> <chr [2]> csat_top
#> 9 All All both_plus_interaction <lgl [1]> <chr [1]> csat
#> 10 All All both_plus_interaction <lgl [1]> <chr [1]> csat_top
#> # … with 70 more rows
Next, we update the formula, generate the data on the fly, calculate our model and prepare the results using ‘broom’. Note that we do all of this within just one call to
dplyr::mutate(), since it allows us to define multiple columns and append them to our nested data.frame. Finally, we drop columns that we don’t need anymore.
all_grps_grid_final_res <- all_grps_grid_final |>
rowwise() |>
mutate(
# dynamically name list
form = list2( "{product}_{type}_{model_spec}_{dep_vars}" :=
# update formula
update(min_formula, # old formula
reformulate(c(".", update_vars), dep_vars)) # changes to formula
),
mod = list(
lm(form,
# create data on the fly
data = filter(csat_named_top,
.env$product == "All" | .env$product == product,
eval(filter_ls)
)
)
),
res = list(broom::tidy(mod)),
modstat = list(broom::glance(mod))
) |>
select(product:model_spec, dep_vars, mod:modstat)
all_grps_grid_final_res
#> # A tibble: 80 × 7
#> # Rowwise:
#> product type model_spec dep_vars mod res modstat
#> <chr> <chr> <chr> <chr> <list> <list> <list>
#> 1 All All base csat <lm> <tibble [4 × 5]> <tibble>
#> 2 All All base csat_top <lm> <tibble [4 × 5]> <tibble>
#> 3 All All email csat <lm> <tibble [5 × 5]> <tibble>
#> 4 All All email csat_top <lm> <tibble [5 × 5]> <tibble>
#> 5 All All website csat <lm> <tibble [5 × 5]> <tibble>
#> 6 All All website csat_top <lm> <tibble [5 × 5]> <tibble>
#> 7 All All both csat <lm> <tibble [6 × 5]> <tibble>
#> 8 All All both csat_top <lm> <tibble [6 × 5]> <tibble>
#> 9 All All both_plus_interaction csat <lm> <tibble [7 × 5]> <tibble>
#> 10 All All both_plus_interaction csat_top <lm> <tibble [7 × 5]> <tibble>
#> # … with 70 more rows
As we add more complex functions to our workflow, we can see that the code gets pretty messy, and hard to debug. To keep the code clean and to allow for easier debugging we can tuck away the more complex parts of our code in customs functions. More details on this can be found in the info box below.
Expand: Use custom functions for cleaner code
In the code above our mutate call was spanning more than twenty lines of code. This makes it not only hard to see what’s actually going on, it also makes debugging much more difficult.
In this case it is helpful to tuck the more complex parts of our code away in custom function. Taking our last example, we could create two functions update_min_form(), that updates the model formula, and lineaer_mod(), that calculates a linear model and creates the data on th fly.
# define custom functions
update_min_form <- function(upvars, dpvars) { # update_vars, dep_vars
update(min_formula, # old formula
reformulate(c(".", upvars), dpvars) # changes to formula
)
}
linear_mod <- function(form, prodvar, filter_inp) {
lm(form,
# create data on the fly
data = filter(csat_named_top,
prodvar == "All" | .data$product == prodvar,
eval(filter_inp)
)
)
}With both functions predefined, our original code becomes much clearer to read and easier to debug:
all_grps_grid_final_res2 <- all_grps_grid_final |>
rowwise() |>
mutate(
form = list2("{product}_{type}_{model_spec}_{dep_vars}" :=
update_min_form(update_vars, dep_vars)
),
mod = list(linear_mod(form, product, filter_ls)),
res = list(broom::tidy(mod)),
modstat = list(broom::glance(mod))
) |>
select(product:model_spec, dep_vars, mod:modstat)
all_grps_grid_final_res2
#> # A tibble: 80 × 7
#> # Rowwise:
#> product type model_spec dep_vars mod res modstat
#> <chr> <chr> <chr> <chr> <list> <list> <list>
#> 1 All All base csat <lm> <tibble [4 × 5]> <tibble>
#> 2 All All base csat_top <lm> <tibble [4 × 5]> <tibble>
#> 3 All All email csat <lm> <tibble [5 × 5]> <tibble>
#> 4 All All email csat_top <lm> <tibble [5 × 5]> <tibble>
#> 5 All All website csat <lm> <tibble [5 × 5]> <tibble>
#> 6 All All website csat_top <lm> <tibble [5 × 5]> <tibble>
#> 7 All All both csat <lm> <tibble [6 × 5]> <tibble>
#> 8 All All both csat_top <lm> <tibble [6 × 5]> <tibble>
#> 9 All All both_plus_interaction csat <lm> <tibble [7 × 5]> <tibble>
#> 10 All All both_plus_interaction csat_top <lm> <tibble [7 × 5]> <tibble>
#> # … with 70 more rows
Although we are not specifically interested in the results, below is one way to filter out those models that are statistically significant at the 10% level arranged by adjusted r-squared in descending order:
all_grps_grid_final_res |>
unnest(modstat) |>
select(-c(sigma, statistic, df:df.residual)) |>
filter(p.value < 0.1) |>
arrange(desc(adj.r.squared))
#> # A tibble: 4 × 10
#> product type model…¹ dep_v…² mod res r.squ…³ adj.r…⁴ p.value nobs
#> <chr> <chr> <chr> <chr> <lis> <list> <dbl> <dbl> <dbl> <int>
#> 1 advanced no_reac… base csat_t… <lm> <tibble> 0.423 0.279 0.0765 16
#> 2 advanced All base csat_t… <lm> <tibble> 0.349 0.234 0.0575 21
#> 3 All All email csat <lm> <tibble> 0.112 0.0575 0.0973 70
#> 4 All All base csat <lm> <tibble> 0.0884 0.0538 0.0613 83
#> # … with abbreviated variable names ¹model_spec, ²dep_vars, ³r.squared,
#> # ⁴adj.r.squared
Wrap-up
That’s it! This post started out easy and got quite complex in the end. I hope to have shown how the original Many Models Approach can be refined from running the same model on various distinct subsets of data to running different variations of a model on many partly overlapping subsets. I hope you enjoyed this post. If you use other helper functions or have a better approach for calculating many models, I’d love to hear your feedback. Feel free to share your thoughts in the comments below or via Twitter, Mastodon, or GitHub.
Acknowledgements
I’m thankful for the many comments I have received for the initial version of this blog post. Isabelle Ghement was so kind to provide in-depth feedback which helped me to, hopefully, explain some of the concepts more concisely. Many thanks for that!
Session Info
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.2.1 (2022-06-23)
#> os macOS Big Sur ... 10.16
#> system x86_64, darwin17.0
#> ui X11
#> language (EN)
#> collate en_US.UTF-8
#> ctype en_US.UTF-8
#> tz Europe/Berlin
#> date 2023-07-12
#> pandoc 2.19.2 @ /Applications/RStudio.app/Contents/MacOS/quarto/bin/tools/ (via rmarkdown)
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> backports 1.4.1 2021-12-13 [1] CRAN (R 4.2.0)
#> broom * 1.0.1 2022-08-29 [1] CRAN (R 4.2.0)
#> cachem 1.0.6 2021-08-19 [1] CRAN (R 4.2.0)
#> cli 3.6.0 2023-01-09 [1] CRAN (R 4.2.0)
#> digest 0.6.31 2022-12-11 [1] CRAN (R 4.2.0)
#> downlit 0.4.2 2022-07-05 [1] CRAN (R 4.2.0)
#> dplyover * 0.0.8.9002 2022-10-15 [1] Github (timteafan/dplyover@f0cd984)
#> dplyr * 1.1.0 2023-01-29 [1] CRAN (R 4.2.0)
#> ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.2.0)
#> evaluate 0.19 2022-12-13 [1] CRAN (R 4.2.0)
#> fansi 1.0.3 2022-03-24 [1] CRAN (R 4.2.0)
#> fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.2.0)
#> fs 1.5.2 2021-12-08 [1] CRAN (R 4.2.0)
#> generics 0.1.3 2022-07-05 [1] CRAN (R 4.2.0)
#> glue 1.6.2 2022-02-24 [1] CRAN (R 4.2.0)
#> highr 0.10 2022-12-22 [1] CRAN (R 4.2.0)
#> htmltools 0.5.4 2022-12-07 [1] CRAN (R 4.2.0)
#> hugodownplus 0.0.0.9000 2023-02-19 [1] Github (timteafan/hugodownplus@d79c4c0)
#> knitr 1.41 2022-11-18 [1] CRAN (R 4.2.0)
#> lifecycle 1.0.3 2022-10-07 [1] CRAN (R 4.2.0)
#> magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.2.0)
#> memoise 2.0.1 2021-11-26 [1] CRAN (R 4.2.0)
#> modelsummary * 1.3.0 2023-01-05 [1] CRAN (R 4.2.0)
#> openxlsx 4.2.5.2 2023-02-06 [1] CRAN (R 4.2.0)
#> pillar 1.8.1 2022-08-19 [1] CRAN (R 4.2.0)
#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.2.0)
#> purrr * 1.0.1 2023-01-10 [1] CRAN (R 4.2.0)
#> R6 2.5.1 2021-08-19 [1] CRAN (R 4.2.0)
#> Rcpp 1.0.9 2022-07-08 [1] CRAN (R 4.2.0)
#> rlang * 1.0.6 2022-09-24 [1] CRAN (R 4.2.0)
#> rmarkdown 2.19 2022-12-15 [1] CRAN (R 4.2.0)
#> rstudioapi 0.14 2022-08-22 [1] CRAN (R 4.2.0)
#> sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.2.0)
#> stringi 1.7.8 2022-07-11 [1] CRAN (R 4.2.0)
#> stringr 1.5.0 2022-12-02 [1] CRAN (R 4.2.0)
#> tables 0.9.10 2022-10-17 [1] CRAN (R 4.2.0)
#> tibble 3.1.8 2022-07-22 [1] CRAN (R 4.2.0)
#> tidyr * 1.2.1 2022-09-08 [1] CRAN (R 4.2.0)
#> tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.2.0)
#> utf8 1.2.2 2021-07-24 [1] CRAN (R 4.2.0)
#> vctrs 0.5.2 2023-01-23 [1] CRAN (R 4.2.0)
#> withr 2.5.0 2022-03-03 [1] CRAN (R 4.2.0)
#> xfun 0.36 2022-12-21 [1] CRAN (R 4.2.0)
#> yaml 2.3.6 2022-10-18 [1] CRAN (R 4.2.0)
#> zip 2.2.2 2022-10-26 [1] CRAN (R 4.2.0)
#>
#> [1] /Library/Frameworks/R.framework/Versions/4.2/Resources/library
#>
#> ──────────────────────────────────────────────────────────────────────────────