Partially renaming columns using a lookup table
Intro
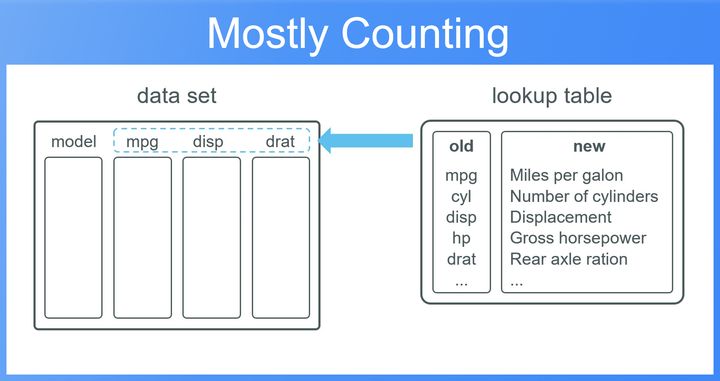
Usually data sets come with short column names, which makes it easy to clean and manipulate the data. However, when presenting the data to stakeholders, in form of tables or plots, we often need longer, meaningful names. In many cases we have a lookup table which contains long and short versions of the column names so that we can “easily” replace the names when needed.
Below we’ll look at how to rename columns using different approaches in R. To make this a little bit more challenging, we’ll add three conditions:
- The lookup table is not complete, that means the lookup table only covers a subset of the columns in our data set.
- We are working with a subset of the original data, that means, the lookup table, although being not complete, holds actually more column name pairs than there are actually columns in the subset of our data.
- The sorting of the lookup table is different from the sorting of our actual column names.
Without those three conditions partially renaming columns is actually not a big deal. In real world settings however, there are many cases where we have to rename columns under one or more of the above conditions. Especially, since we often use short column names in the analysis and just rename them in the final step when creating a report. The latter almost never contains all the columns names of our originial data set.
It is interesting to see how the three large paradigms in R, base R, ‘data.table’ and ‘dplyr’ compare in handling this problem.
This post concludes by looking at how we would tackle the same problem in Python’s ‘pandas’ library.
Let’s start with the setup.
Setup
We take the mtcars data set and create lookup data.frame called recode_df based on the information from the documentation
?mtcars. Next, we apply the three conditions mentioned above (see code comments) and assign this new data to mycars.
recode_df <- data.frame(
old = names(mtcars),
new = c("Miles per galon", "Number of cylinders", "Displacement (cu.in.)",
"Gross horsepower","Rear axle ratio", "Weight (1000 lbs)",
"1/4 mile time", "Engine (0=automatic, 1=manual)",
"Number of forward gears", "Transmission (0=automatic, 1=manual)",
"Number of carbuertors")
)
# condition 3: The lookup table has a different sorting than the actual column names
# Here we choose a alphabetical ordering for the lookup table:
recode_df <- recode_df[order(recode_df$old),]
rownames(recode_df) <- NULL
# condition 2: we are only working with a subset of the data
# Here we only use every second column:
every_2nd_col <- seq(from = 1, to = length(mtcars), by = 2)
mycars <- mtcars[,every_2nd_col]
# condition 1: the data has a column that is not part of the lookup table
# Here we take the rownames and put them in a dedicated column `model` ...
# ... which is no included in `recode_df`
mycars <- cbind(model = rownames(mycars),
data.frame(mycars, row.names = NULL))
str(mycars)
#> 'data.frame': 32 obs. of 7 variables:
#> $ model: chr "Mazda RX4" "Mazda RX4 Wag" "Datsun 710" "Hornet 4 Drive" ...
#> $ mpg : num 21 21 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 ...
#> $ disp : num 160 160 108 258 360 ...
#> $ drat : num 3.9 3.9 3.85 3.08 3.15 2.76 3.21 3.69 3.92 3.92 ...
#> $ qsec : num 16.5 17 18.6 19.4 17 ...
#> $ am : num 1 1 1 0 0 0 0 0 0 0 ...
#> $ carb : num 4 4 1 1 2 1 4 2 2 4 ...
As as final step lets write both data.frame’s mycars and recode_df from R to two separate csv files, so that we can load them easily into Python later on. (In RMarkdown we could of course access the objects created in R from Python via the r object, but lets stick to csv files to make this reproducible for all users.)
write.csv(mycars, "mycars.csv")
# available at:
# read.csv("https://raw.githubusercontent.com/TimTeaFan/tt_website/main/content/post/2022-rename-columns/mycars.csv")
write.csv(recode_df, "recode_df.csv")
# available at:
# read.csv("https://raw.githubusercontent.com/TimTeaFan/tt_website/main/content/post/2022-rename-columns/recode_df.csv")base R
Lets start with base R. If it weren’t for the three conditions outlined above, renaming columns in base R would be really easy. It would basically boil down to a classic lookup using
match() as index to extract [ the new names:
recode_df$new[match(names(mycars), recode_df$old)]
#> [1] NA "Miles per galon"
#> [3] "Displacement (cu.in.)" "Rear axle ratio"
#> [5] "1/4 mile time" "Number of forward gears"
#> [7] "Number of carbuertors"
But as we can see, this creates an NA for the column name that is not included in the lookup table model.
However, slightly modifying
this answer on SO by Gregor Thomas, shows how to get rid of the NAs and only overwrite those column names that are present in our lookup table:
# create a new object so that we don't overwrite our original `mycars` data
mycars_base <- mycars
# create an index vector with match to find those names in`recode_df` ...
# ...that are present in our data
idx_vec <- match(recode_df$old, names(mycars_base))
# assign the names
names(mycars_base)[na.omit(idx_vec)] <- recode_df$new[!is.na(idx_vec)]
# use `str()` for better printing
str(mycars_base)
#> 'data.frame': 32 obs. of 7 variables:
#> $ model : chr "Mazda RX4" "Mazda RX4 Wag" "Datsun 710" "Hornet 4 Drive" ...
#> $ Miles per galon : num 21 21 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 ...
#> $ Displacement (cu.in.) : num 160 160 108 258 360 ...
#> $ Rear axle ratio : num 3.9 3.9 3.85 3.08 3.15 2.76 3.21 3.69 3.92 3.92 ...
#> $ 1/4 mile time : num 16.5 17 18.6 19.4 17 ...
#> $ Number of forward gears: num 1 1 1 0 0 0 0 0 0 0 ...
#> $ Number of carbuertors : num 4 4 1 1 2 1 4 2 2 4 ...
Although this approach is not very verbose, it does require some serious thinking about matching, extracting and indexing. It feels like there should be a cleaner solution for a common problem like this, so lets have a look how we can tackle this problem using ‘data.table’ and ‘dplyr’.
data․table
The ‘data.table’ package sometimes has the reputation for offering a cryptic, arcane syntax, but many users don’t know that the package also contains many helpful functions which are pretty straight-forward to use. In our case we can apply
data.table::setnames() out of the box. It takes a data.table, a vector of old and new column names and finally all we have to do is to set the skip_absent argument to TRUE, to prevent ‘data.table’ from raising an error, since not all of the names in our lookup table are present in the data.
Unlike base R, the names are changed “by reference”, meaning that we don’t need to assign the result to a new variable, since no copy is made. Instead the data is “modified in place”.
library(data.table)
mycars_dt <- as.data.table(mycars)
setnames(mycars_dt,
old = recode_df$old,
new = recode_df$new,
skip_absent = TRUE)
str(mycars_dt)
#> Classes 'data.table' and 'data.frame': 32 obs. of 7 variables:
#> $ model : chr "Mazda RX4" "Mazda RX4 Wag" "Datsun 710" "Hornet 4 Drive" ...
#> $ Miles per galon : num 21 21 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 ...
#> $ Displacement (cu.in.) : num 160 160 108 258 360 ...
#> $ Rear axle ratio : num 3.9 3.9 3.85 3.08 3.15 2.76 3.21 3.69 3.92 3.92 ...
#> $ 1/4 mile time : num 16.5 17 18.6 19.4 17 ...
#> $ Number of forward gears: num 1 1 1 0 0 0 0 0 0 0 ...
#> $ Number of carbuertors : num 4 4 1 1 2 1 4 2 2 4 ...
#> - attr(*, ".internal.selfref")=<externalptr>
dplyr (tidyverse)
Renaming columns in ‘dplyr’ is as easy as df %>% rename("new_name" = "old_name"). At least when we use
dplyr::rename() interactively and type in each old and new name manually. Usually when working programmatically with ‘dplyr’ we supply a named vector to functions that take the ellipsis ... as argument, like
rename(), and splice it in using the bang-bang-bang operator !!!.
library(dplyr)
# create a named vector
recode_vec <- setNames(recode_df$old, recode_df$new)
# splice it in with `!!!`
mycars %>%
rename(!!! recode_vec)
#> Error in `rename()`:
#> ! Can't rename columns that don't exist.
#> ✖ Column `cyl` doesn't exist.
However, this won’t work in our case, since our lookup table, and the named vector that we constructed with it, contains column name pairs that don’t exist in our data, which leads to the above error.
Intuitively, one want’s fall back to the base R approach where we extract only those names from our named vector that our data actually contains:
# extract column name pairs that are actually in our data and splice into `rename()`
mycars %>%
rename(!!! recode_vec[recode_vec %in% names(mycars)]) %>%
glimpse() # for better printing
#> Rows: 32
#> Columns: 7
#> $ model <chr> "Mazda RX4", "Mazda RX4 Wag", "Datsun 710", …
#> $ `Miles per galon` <dbl> 21.0, 21.0, 22.8, 21.4, 18.7, 18.1, 14.3, 24…
#> $ `Displacement (cu.in.)` <dbl> 160.0, 160.0, 108.0, 258.0, 360.0, 225.0, 36…
#> $ `Rear axle ratio` <dbl> 3.90, 3.90, 3.85, 3.08, 3.15, 2.76, 3.21, 3.…
#> $ `1/4 mile time` <dbl> 16.46, 17.02, 18.61, 19.44, 17.02, 20.22, 15…
#> $ `Number of forward gears` <dbl> 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ `Number of carbuertors` <dbl> 4, 4, 1, 1, 2, 1, 4, 2, 2, 4, 4, 3, 3, 3, 4,…
And although this works, it is not as easy and clean as we expect. However, there is an even easier way in ‘dplyr’ which doesn’t come with the need of neither splicing nor extracting:
dplyr::any_of().
Usually
any_of() takes a character vector with column names and is used inside
dplyr::select() to select “any of” the columns in the vector. It won’t throw an error when any of the column names is not actually in our data.
The cool, and undocumented feature is that we can use
any_of() inside
rename() and that we can supply it a named vector to do the renaming for us:
# first lets create new object
# ... so that we don't overwrite our original `mycars` data
mycars_tidy <- mycars
# here we construct the same named vector as above
recode_vec <- setNames(recode_df$old, recode_df$new)
mycars_tidy <- mycars_tidy %>%
rename(any_of(recode_vec))
# use `glimpse()` for better printing
mycars_tidy %>%
glimpse()
#> Rows: 32
#> Columns: 7
#> $ model <chr> "Mazda RX4", "Mazda RX4 Wag", "Datsun 710", …
#> $ `Miles per galon` <dbl> 21.0, 21.0, 22.8, 21.4, 18.7, 18.1, 14.3, 24…
#> $ `Displacement (cu.in.)` <dbl> 160.0, 160.0, 108.0, 258.0, 360.0, 225.0, 36…
#> $ `Rear axle ratio` <dbl> 3.90, 3.90, 3.85, 3.08, 3.15, 2.76, 3.21, 3.…
#> $ `1/4 mile time` <dbl> 16.46, 17.02, 18.61, 19.44, 17.02, 20.22, 15…
#> $ `Number of forward gears` <dbl> 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ `Number of carbuertors` <dbl> 4, 4, 1, 1, 2, 1, 4, 2, 2, 4, 4, 3, 3, 3, 4,…
The good thing about both approaches in ‘data.table’ and ‘dplyr’ is that we can deliberately ignore errors when not all of the column name pairs are present in our data. Either by setting
data.table::setnames()‘s skip_absent argument to TRUE or by using
tidyselect::any_of(). Which means we can also raise errors if we need them: setting skip_absent to FALSE or using
tidyselect::all_of() (which is the opposite of
any_of() and requires all column names to be present in the data).
Finally, lets have a look at how we would solve this problem in Python’s ‘pandas’ library.
Python pandas
In Python we can use ‘pandas’s
pd.DataFrame.rename function to rename columns of a DataFrame. The only thing we need to take care of is supplying the columns argument with a dictionary of column name pairs {"old" : "new"}. There are several ways to create a dictionary from our recode_df. Below we use a pandas approach with set_index(...).to_dict(), but we could also have used Pythons dict(zip()) functions.
import pandas as pd
# setup
mycars = pd.read_csv("mycars.csv", index_col = 0)
recode_df = pd.read_csv("recode_df.csv", index_col = 0)
# Create dictionary from `recode_df`
recode_dic = recode_df.set_index('old')['new'].to_dict()
# Alternatively: dict(zip(recode_df['old'], recode_df['new']))
# Rename columns
(mycars
.rename(
columns=recode_dic,
inplace=True
)
)
mycars.info()
#>
#> Int64Index: 32 entries, 1 to 32
#> Data columns (total 7 columns):
#> # Column Non-Null Count Dtype
#> --- ------ -------------- -----
#> 0 model 32 non-null object
#> 1 Miles per galon 32 non-null float64
#> 2 Displacement (cu.in.) 32 non-null float64
#> 3 Rear axle ratio 32 non-null float64
#> 4 1/4 mile time 32 non-null float64
#> 5 Number of forward gears 32 non-null int64
#> 6 Number of carbuertors 32 non-null int64
#> dtypes: float64(4), int64(2), object(1)
#> memory usage: 2.0+ KB
When it comes to renaming columns, we can see that ‘pandas’ is pretty similar to ‘dplyr’, even more so when we write our Python code according to the "Effective Pandas" style. However, it also resembles ‘data.table’ in two aspects. First, when setting the inplace argument to True the DataFrame is modified in place, no copy is made, and we don’t need to assign the result back to a variable. Second, rename has an argument errors which is set to 'ignore' by default. If we want pandas to throw an error if not all columns are present in our data, we can set it to 'raise'.
That’s it. I hope you enjoyed reading about renaming columns in R and Python. If you have a better way of renaming columns (especially in base R) let me know in the comments below or via Twitter, Mastodon or Github.
Session Info
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.2.1 (2022-06-23)
#> os macOS Big Sur ... 10.16
#> system x86_64, darwin17.0
#> ui X11
#> language (EN)
#> collate en_US.UTF-8
#> ctype en_US.UTF-8
#> tz Europe/Berlin
#> date 2023-02-05
#> pandoc 2.19.2 @ /Applications/RStudio.app/Contents/MacOS/quarto/bin/tools/ (via rmarkdown)
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> data.table * 1.14.2 2021-09-27 [1] CRAN (R 4.2.0)
#> dplyr * 1.1.0 2023-01-29 [1] CRAN (R 4.2.0)
#> reticulate * 1.26 2022-08-31 [1] CRAN (R 4.2.0)
#>
#> [1] /Library/Frameworks/R.framework/Versions/4.2/Resources/library
#>
#> ─ Python configuration ───────────────────────────────────────────────────────
#> python: /usr/local/Caskroom/miniconda/base/bin/python3.9
#> libpython: /usr/local/Caskroom/miniconda/base/lib/libpython3.9.dylib
#> pythonhome: /usr/local/Caskroom/miniconda/base:/usr/local/Caskroom/miniconda/base
#> version: 3.9.12 (main, Apr 5 2022, 01:53:17) [Clang 12.0.0 ]
#> numpy: /usr/local/Caskroom/miniconda/base/lib/python3.9/site-packages/numpy
#> numpy_version: 1.22.3
#>
#> NOTE: Python version was forced by RETICULATE_PYTHON
#>
#> ──────────────────────────────────────────────────────────────────────────────